Designing Gene Targeting Vectors
Introduction
The generation of mutant mice by gene targeting takes advantage of the remarkable ability of embryonic stem (ES) cell lines (1, 2) to participate in the formation of germ cells of mice when the cells are put back into an early embryo (3). Cell lines which have undergone gene targeting are enriched by the incorporation of selectable markers into the targeting vector (4). Out of this enriched set of cell lines, the desired homologous recombination event is identified by molecular analysis of genomic DNA. Targeted ES cell lines which have a normal number of chromosomes are identified and selected to make chimeras.
Targeting can be used to generate knockouts, knockins or conditional alleles. Knockouts, knockins and conditional alleles are generated in a similar fashion using the protocols given here. Knockouts are used to define the overall requirement for a gene and to model loss of function mutations. Knockins introduce heterologous coding sequences such as reporters or DNA recombinases, or to incorporate changes in DNA sequence to "humanize" a mouse gene or to generate point mutations. Conditional mutations are used to define the organ, tissue or cellular autonomy of mutant effects, to circumvent embryonic lethality, or to model somatic mutations.
The two major hurdles for the generation of gene targeted mice are obtaining homologous recombination and germ line transmission. Although it has been almost twenty years since the first gene targeted mice were constructed (4, 5), not all parameters which affect the frequency of homologous recombination and germ line transmission are known. Nonetheless, we give criteria for the parameters which are known, provide advice on how to ensure success, and provide recovery strategies and trouble shooting guidance.
Targeting Vector Design: General Principles
Our purpose is to present the most common and widely applicable vector designs (Figure 1). There are a large number of variations possible in the design of targeting vectors and the uses they are put to (7,8). The following concentrates on the most common applications.
For all targeting vectors, the following considerations apply: 1) the vector needs to be linearized outside of the arms of homology, so provision must be made for a unique recognition sequence for a restriction enzyme at an appropriate place; 2) a strategy to detect gene targeting by Southern blot analysis of genomic DNA must be developed; 3) the greater the amount of sequence match, the more likely the targeting is to succeed.
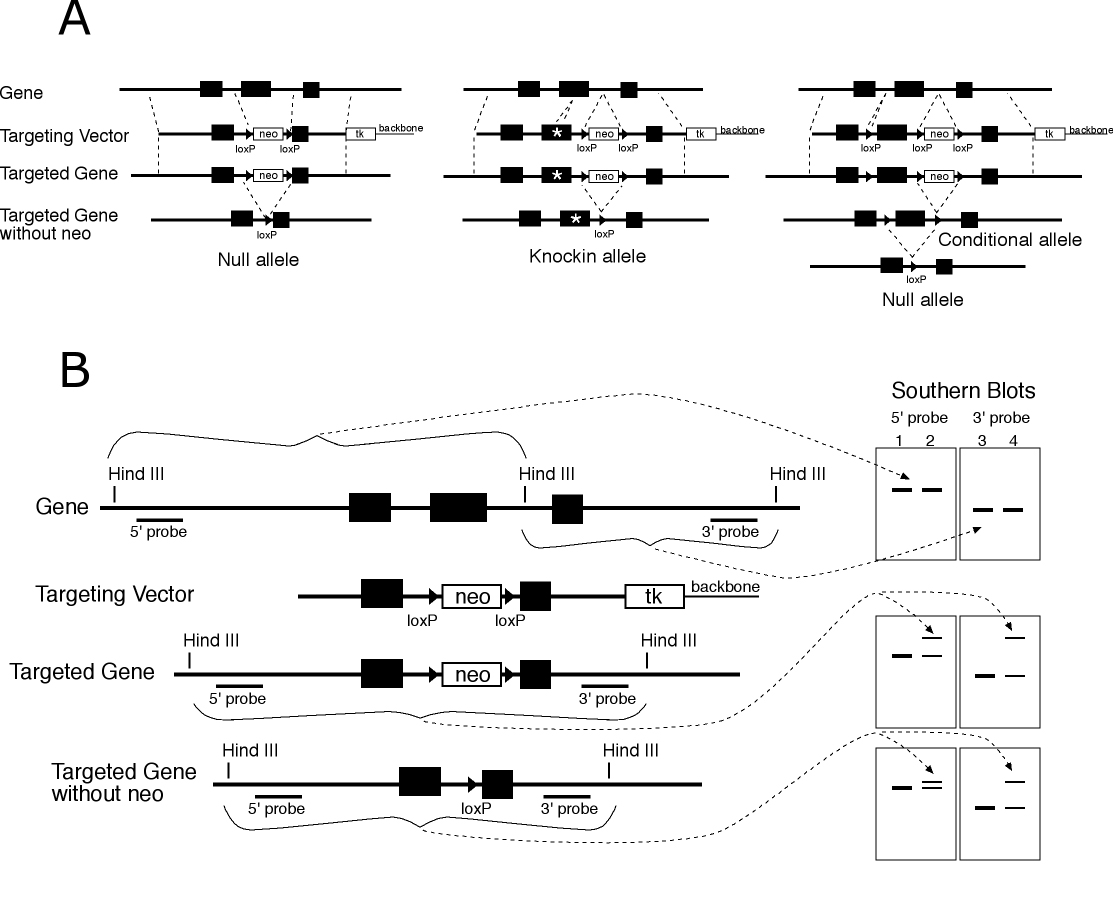
Figure 1 Vector design and detection of targeting. A) Vector designs for gene targeted null, knockin and conditional alleles are shown (left to right). The wild type allele with three exons is shown at top, the targeting vector on the next line, the correctly targeted allele on the next line and the targeted allele after excision of the neo cassette by transfection of Cre recombinase in ES cells. B) Strategy for detection of correctly targeted alleles. DNA probes for Southern blot analysis are selected which are in the gene to be targeted, but external to the targeting vector (5' and 3' probes). After targeting, random insertions of the vector which survive the selection do not show alteration of the endogenous gene (lanes 1 and 3) but correctly targeted lines show a wild type allele and the predicted fragment for a targeted allele. Excision of neo is also verified by observing the predicted changes in fragment size. The detection strategy shown is based on deletion of a Hind III site and change in size of fragments, but a strategies can also be based on introduction of sites. (neo and tk indicate the minigenes consisting of promoters, coding sequences and polyadenylation signals for the neomycin resistance and HSV thymidine kinase genes respectively.)
Targeting Vectors for Null Alleles
We recommend that a targeting vector for construction of a null allele be constructed with genomic DNA from the 129 strain of mice, with at least 7 kb of total homology split in two arms which are positioned to delete early or critical coding exons of the gene (Figure 1). Two selectable markers are incorporated into the vector (4). The neomycin resistance gene (neo) should be flanked by loxP sites, and located between the two arms. The herpes simplex virus (HSV) thymidine kinase (tk) gene, should be placed outside one of the arms of the vector. The neomycin resistance gene is removed in ES cells through the activity of Cre recombinase acting on the loxP sites after gene targeting.
Targeting Vectors for Knockin Alleles
To create a knockin allele (Figure 1), a sequence change or inserted coding sequence is incorporated into one of the arms of the targeting vector (9). The goal is to minimize other disruption of gene function. The neomycin resistance gene is flanked by loxP sites so that it can be excised by expression of Cre recombinase and is typically inserted into an intron. In designing knockins, particularly for those generating small or subtle changes, provision needs to be made to detect if the knockin change itself has been integrated as was intended--this is necessary because a homologous recombination exchange could occur internal to the intended change and not incorporate the altered sequence (see the example in Figure 2 for a conditional allele: the same applies to knockin alleles). The neo marker is removed in ES cells before constructing chimeras.
Targeting Vectors for Conditional Alleles
A conditional allele has wild type function but can be mutated to a null allele in cells in which Cre recombinase is expressed (10). To maintain wild type function, no part of the gene is deleted, and loxP sites are inserted into introns away from sequences which might function in splicing and transcription (Figure 1).
Targeting vectors for conditional alleles should be constructed as for null alleles, except that the neomycin resistance gene with its flanking loxP sites is inserted into an intron, and a loxP is inserted into different intron, such that deletion of sequences between the most distal loxP sites will remove an essential coding exon.
Similar to Knockin strategies, provision must be made to detect incorporation of the distal loxP site, since not all homologous recombinants will have incorporated the loxP at a distance from neo (Figure 2)
The neomycin resistance gene is removed with Cre recombinase in ES cells, and alleles with the two loxP sites flanking the essential exon are identified. The conditional allele is introduced into mice through chimeras, and the gene is mutated to a null allele through the action of Cre recombinase on the two remaining loxP sites, deleting the sequences between them.
It is important to verify that the loxP sequences will be functional and that no unintended sequence alterations were introduced. Therefore, it is wise to sequence the entire targeting vector for conditional alleles before committing to targeting. This is especially the case if parts of the vector have been constructed by PCR.
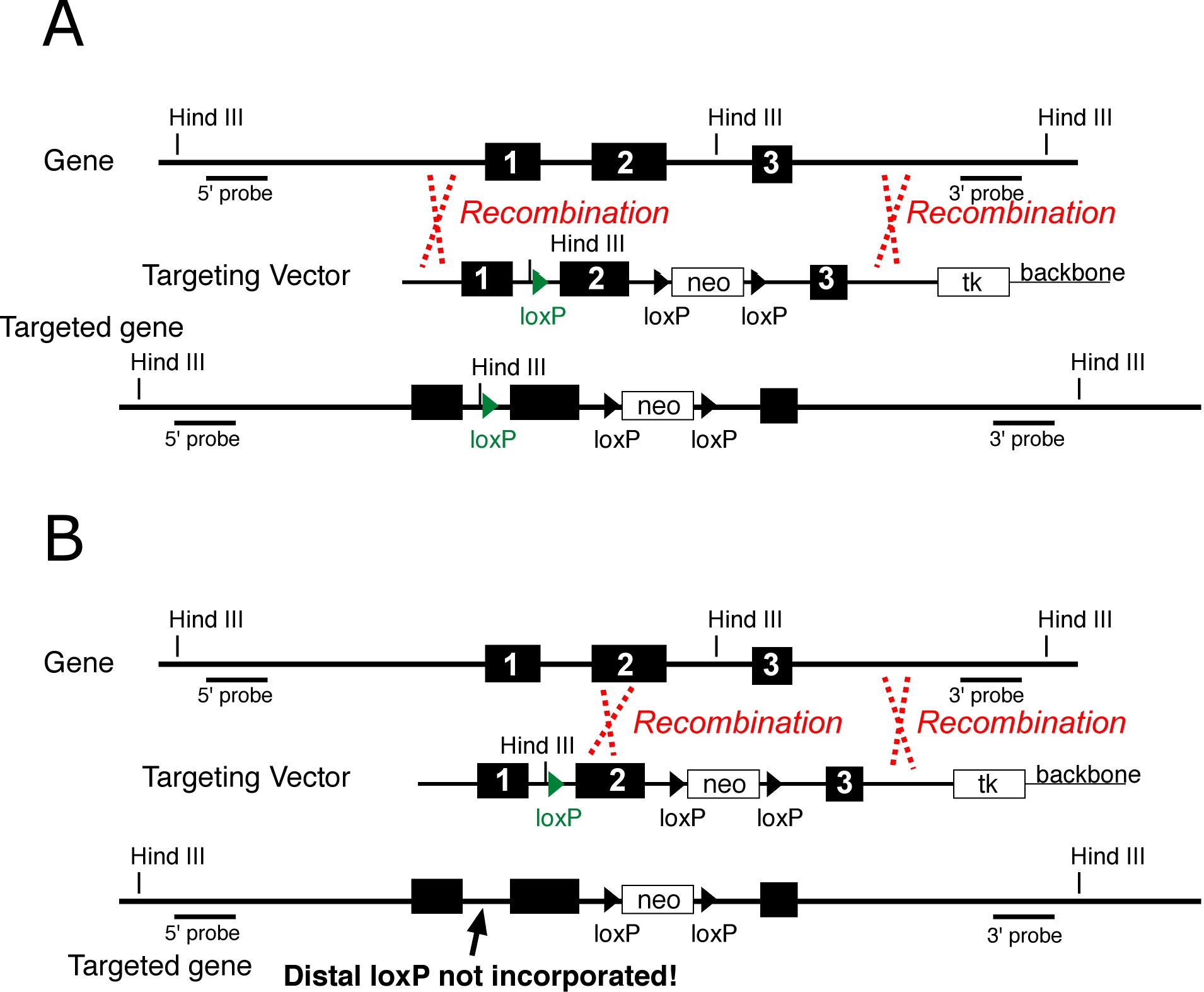
Figure 2 The placement of crossovers (red dotted lines) can result in the inclusion (A) or exclusion (B) of linked sequence alterations, in this case a distal loxP site (green). A targeting vector to construct a conditional allele is shown. The distal loxP (green) has an introduced Hind III site to facilitate detection, and the floxed neo deletes a Hind III site, also to facilitate detection. The same principles apply to knockin alleles where the sequence alteration is separated from neo by homologous DNA.
Isogenic DNA
The frequency of homologous recombination depends on the degree of sequence match and the length of the matching sequences (11, 12). Greater than 7 kilobases of DNA of perfect sequence match usually is needed to obtain homologous recombination at practical frequencies. The relationship between sequence divergence and targeting rates has not been examined systematically, but about 0.5% sequence divergence was shown to result in a 20-fold decrease in targeting frequency for constructs targeting the Rb locus (11). In a targeting vector, the exact sequence match typically is interrupted by a gap and/or an insertion with each of the two parts of the match approximately halved. The apparent paradox that the sequence match with the target gene must be near exact whereas the gap or insertion can be as large as 10 kb probably arises because there are two independent homologous recombination events which occur in gene targeting, one for each arm.
The genomic sequences of different inbred strains of mice diverge enough that DNA from the same strain as the ES cells likely is needed to construct the targeting vector. Most ES cells are made from the 129 strain of mice, thus targeting vectors typically are made from 129 genomic DNA. There are several different substrains of the 129 strain, but differences between them are unlikely to be great enough to affect gene targeting. Genomic libraries of 129 mice are available in lambda phage vectors from commercial vendors (Stratagene 946313). 129 genomic clones in BAC vectors (.pdf) can be purchased from the Sanger Institute. BACs containing your gene can be identified using the ensembl genome browser, selecting the DAS source "129S7/AB2.2 clones". If you wish to obtain a specific clone, clicking on the BAC will bring up a menu, and selecting the link at the bottom of the list will take you to the order form.
Alternatively, DNA from C57BL/6J BAC genomic clones can be used to construct targeting vectors for a 129 ES cell line if there are no polymorphisms in the region of the gene between C57BL/6J and 129S1/SvImJ strains of mice. The differences between 129 and C57BL/6J sequences are distributed in blocks consisting of divergent and essentially identical sequence. Whether there are single nucleotide polymorphisms between 129 and C57Bl/6 for your genomic region can be ascertained here on the Jax web site, and C57BL6J BAC clones can be identified with the UC Santa Cruz genome browser and purchased from BACPAC Resources Center CHORI.
Alternatively, long range PCR can be used to isolate construct arms from ES cell genomic DNA. DNA recovered by PCR should be sequenced to ensure that PCR did not introduce unintended mutations into the targeting vector, particularly for knockin and conditional gene targeting.
Lastly, a C57BL/6 ES cell line can be targeted with DNA constructed from C57BL/6 (or 129 if there are no polymorphisms in that piece of DNA).
Selectable Markers
Heterologous genes are incorporated into the targeting vector for enrichment of targeting events. Between the arms of the targeting vector a gene consisting of a promoter, coding sequence for a drug resistance protein and a polyadenylation signal is incorporated. The neomycin resistance coding sequence is typically used, as this confers resistance to the neomycin analogue G418. Outside of one arm, a gene to express Herpes Simplex Virus thymidine kinase (tk) is placed. In a successful homologous recombination, the tk gene is not integrated into the genome and is lost. Cells in which tk has integrated into genomic DNA can be killed by selection with the drug FIAU. In most transfections, the majority of cells incorporating the targeting vector do not do so by homologous recombination, so most cells incorporate the tk selectable marker, and the FIAU selection kills these cells. Alternative drug selection genes are available as well (8).
The orientation of the selectable marker gene transcription units relative to each other and the targeted gene do not appear to be important.
Avoiding Unintended Consequences
It is important to anticipate the consequences of gene targeting to ensure that the targeted gene lacks all function as intended. Examine the splicing patterns which might be expected to occur in the targeted allele, looking for in-frame splicing which could result in aberrant protein products with altered activity. Avoid designs which might give rise to proteins with dominant negative or other gain of function activities.
The neomycin resistance coding sequence and its PGK1 promoter can have unintended consequences on the targeted gene and on adjacent genes if they are left in the gene (13-16). The neomycin resistance gene contains cryptic splice acceptors and donors which can be utilized by transcripts from targeted gene. In addition, the function of genes adjacent to the targeted gene can be altered by the neomycin gene inserted at the targeted locus. In one case this interference has been shown to be due to transcription from the PGK1 promoter and aberrant splicing of neo sequences into the adjacent gene, but other mechanisms are possible, including interference through enhancer competition. If the targeting vector is designed with recognition sequences for a site-specific DNA recombinase flanking the neo gene as describe above, it can be removed after gene targeting by expression of the DNA recombinase. The Cre site-specific DNA recombinase recognizes a 34 base pair sequences, the loxP site. Sequences between a pair of matched sites in direct repeat orientation are deleted, leaving a single loxP sequence.
Planning For Detection Of Gene Targeting
A strategy for identifying the targeted locus by Southern blot analysis must be developed in parallel with vector design (Figure 1). Two DNA probes from the gene on each side outside the targeting vector must be able to detect a change in fragment size, resulting either from the introduction or elimination of a restriction enzyme site.
Avoid the use of restriction enzymes which have recognition sequences which have 5'CG3' dinucleotides because this sequence is most often methylated in ES cell genomic DNA and will be resistant to cutting. For example, the recognition sequence for Xho I is 5'CTCGAG3' and thus Xho I should be avoided; Hind III recognizes the sequence 5'AAGCTT3' and is suitable. Before transfecting ES cells with the targeting vector, test the probes for Southern blot analysis on a genomic digest of ES cell DNA to verify that they work well.
It is important to verify the homologous recombination event from both sides with probes external to the targeting vector since recombination can occur on one side only (17-18 and Figure 2). In practice, the initial identification of homologous recombination events is done with one external probe, then the candidate targeted cell lines are thawed, larger amounts of genomic DNA are prepared, and targeting is verified by extensive genomic Southern blot analysis using multiple probes and digests.
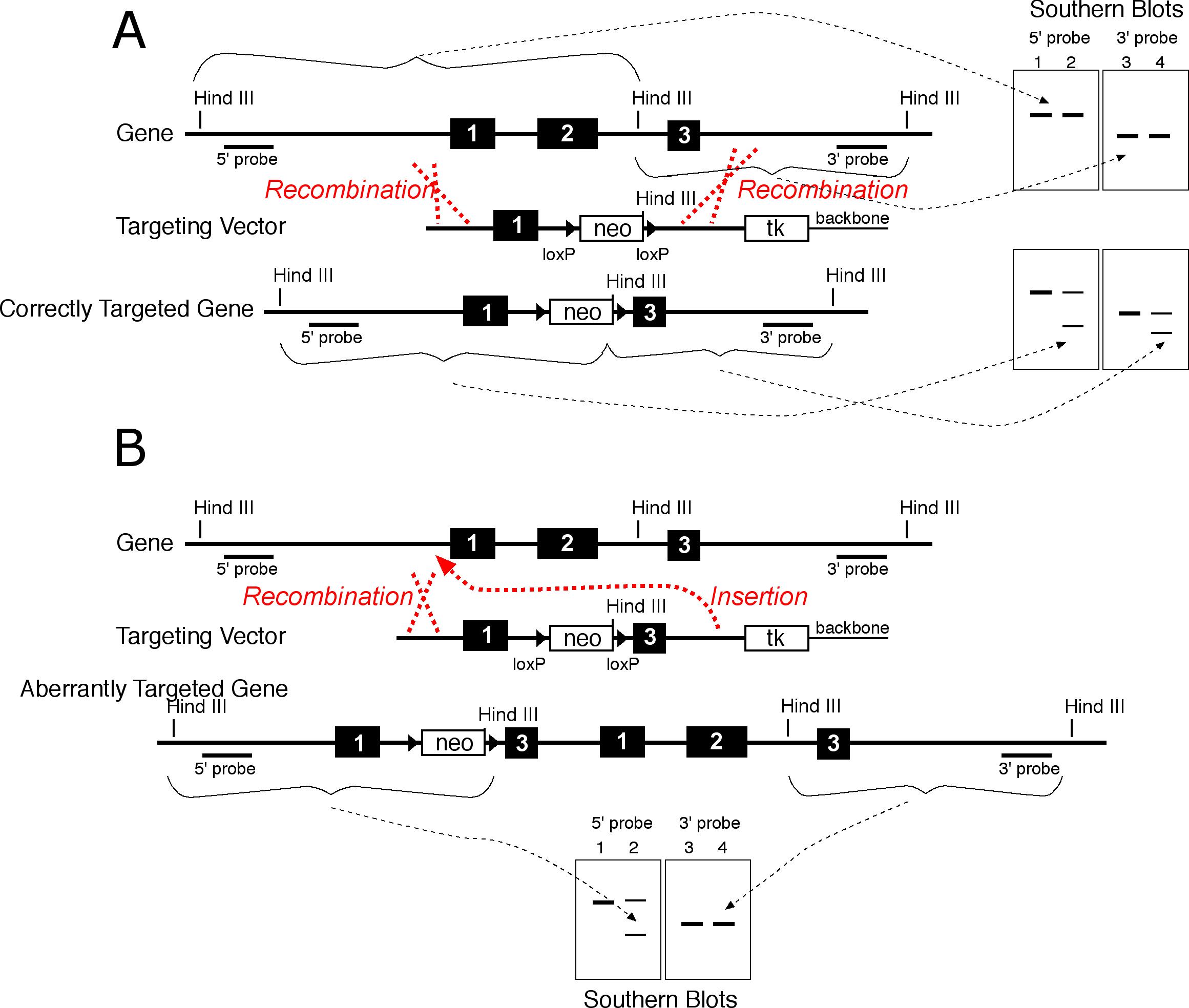
Figure 3 Illustration of correct targeting (A), and aberrant targeting (B). In some cases, homologous recombination occurs on one side only as shown (B). B) Homologous recombination occurred at the 5' end but the 3' end was inserted. In this case, the 5' external probe shows the expected fragments for a targeted allele, but the 3' side shows the wild type fragment. Thus, external probes on both sides should be used to verify that targeting acheived the desired result.
Homologous recombination can occur in ways which can result in products which are not desired. Homologous recombination can occur at one end of the targeting vector with a duplication on the other end. These recombinants are excluded by detecting the expected products with both the 3' external probe and the 5' external probe. The side at which homologous recombination occurred will give the expected, targeted pattern in a Southern blot, but the duplicated side will give a wild type pattern.
In conditional and point mutant targeting strategies, engineered sequence elements (point mutations or loxP sites) at a distance from the neo cassette might not be incorporated. This can occur because recombination took place internally, between the element and neo. Thus, the strategy for detection of conditional and point mutants must incorporate a means to detect elements which are at a distance from neo.
Vector Linearization
Targeting vectors are linearized before transfection, so provision needs to be made for a unique recognition sequence to linearize the vector. Ideally, this site is place such that when the vector is cut, the vector has at one end one arm of homology, and at the other end the vector backbone is external to the tk gene.
Assembling The Targeting Vector
We do not provide a protocol describing construction of the targeting vector, but the technology involved is that common to most molecular biology subcloning. There are companies which will construct targeting vectors for a fee. Plasmids carrying the neo selectable marker flanked by loxP sites, pflox (19), and the tk selectable marker, pPNT (20), can be obtained from the labs they originated in. Contact us for details.
References
- Martin, G. R. (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 78, 7634-8.
- Evans, M. J., and Kaufman, M. H. (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154-6.
- Bradley, A., Evans, M., Kaufman, M. H., and Robertson, E. (1984) Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255-6.
- Thomas, K. R., and Capecchi, M. R. (1987) Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51, 503-12.
- Doetschman, T., Gregg, R. G., Maeda, N., Hooper, M. L., Melton, D. W., Thompson, S., and Smithies, O. (1987) Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330, 576-8.
- Davis, J. (Ed.) (2002) Basic cell culture: a practical approach. Oxford University Press, Oxford University Press, Oxford.
- Nagy, A., Gertsenstein, M., Vintersten, K., and Behringer, R. (2003) Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
- Hasty, P., Abuin, A., and Bradley, A. (2000) Gene targeting, principles, and practice in mammalian cells, in Gene Targeting. A Practical Approach. (Joyner, A. L., Ed.), Vol. 212, Oxford University Press, Oxford, pp. 1-35.
- Hanks, M., Wurst, W., Anson-Cartwright, L., Auerbach, A. B., and Joyner, A. L. (1995) Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science 269, 679-82.
- Gu, H., Marth, J. D., Orban, P. C., Mossmann, H., and Rajewsky, K. (1994) Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 265, 103-6.
- te Riele, H., Maandag, E. R., and Berns, A. (1992) Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci U S A 89, 5128-32.
- Hasty, P., Rivera-Perez, J., and Bradley, A. (1991) The length of homology required for gene targeting in embryonic stem cells. Mol Cell Biol 11, 5586-91.
- Meyers, E. N., Lewandoski, M., and Martin, G. R. (1998) An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet 18, 136-41.
- Nagy, A., Moens, C., Ivanyi, E., Pawling, J., Gertsenstein, M., Hadjantonakis, A. K., Pirity, M., and Rossant, J. (1998) Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr Biol 8, 661-4.
- Olson, E. N., Arnold, H. H., Rigby, P. W., and Wold, B. J. (1996) Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85, 1-4.
- Ren, S. Y., Angrand, P. O., and Rijli, F. M. (2002) Targeted insertion results in a rhombomere 2-specific Hoxa2 knockdown and ectopic activation of Hoxa1 expression. Dev Dyn 225, 305-15.
- Hasty, P., Rivera-Perez, J., Chang, C., and Bradley, A. (1991) Target frequency and integration pattern for insertion and replacement vectors in embryonic stem cells. Mol Cell Biol 11, 4509-17.
- Moens, C. B., Auerbach, A. B., Conlon, R. A., Joyner, A. L., and Rossant, J. (1992) A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev 6, 691-704.
- Chui, D., Oh-Eda, M., Liao, Y. F., Panneerselvam, K., Lal, A., Marek, K. W., Freeze, H. H., Moremen, K. W., Fukuda, M. N., and Marth, J. D. (1997) Alpha-mannosidase-II deficiency results in dyserythropoiesis and unveils an alternate pathway in oligosaccharide biosynthesis. Cell 90, 157-67.
- Tybulewicz, V. L., Crawford, C. E., Jackson, P. K., Bronson, R. T., and Mulligan, R. C. (1991) Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65, 1153-63.